Advance Data Analytics and Visualization using Pathology Test Ordering Data to Optimize Pathology Laboratory Utilization
Collaborators: Calvino Cheng, Manal Elnenaei, Bryan Crocker (Dept. of Pathology and Laboratory Medicine, NSHA), Samuel Campbell (Dept. of Emergency Medicine, NSHA)
Synopsis: The ‘Choosing Wisely Canada’ movement is promoting the provision of sustainable healthcare by optimizing the utilization of healthcare services and adhering to best practices in clinical decision making. It has been observed that a significant number of pathology tests ordered by physicians are inappropriate, leading to suboptimal laboratory utilization and patient safety. In this project, we are employing big’ data analytics, machine learning and data visualization methods to develop a laboratory utilization management platform termed as Pathology Laboratory Utilization Scorecards (PLUS) to provide test ordering insights in terms of (i) interactive scorecards for physicians to examine their test ordering pattern across different tests and over time, rate of inappropriate tests, compliance with clinical guidelines and how they compare with peers, and (ii) interactive dashboards for laboratory managers to assist them with resource planning and waste minimization. PLUS will be implemented to optimize the central zone pathology laboratory in Halifax (the largest provincial lab) that on average processes 8 million general lab orders.
Using Predictive Analytics for Stratification of Surgical Patients in Orthopaedics to Reduce Surgery Wait times and Improve Surgical Outcomes.
Collaborators: Michael Dunbar, Kathryn Young-Shand (Dept. of Orthopaedics, NSHA), Janie Wilson (Dept. of Biomedical Engineering, Dalhousie University)
Synopsis: Reducing the wait lists for Total Knee Arthroplasty (TKA) whilst improving surgical outcomes is a major consideration for the Nova Scotia healthcare system, and even across Canada. One approach to improve surgical outcomes is to objectively select TKA recipients with the greatest need and improvement potential. To better screen candidate patients, we are investigating how a combination of preoperative and postoperative clinical and biomechanical features correlate with objective surgical outcomes. The predictive analytics aspect of this research involves the use of machine learning methods to (a) identify relevant patient phenotypes and clinical biomechanical phenotypes among a heterogeneous group of TKA candidates with differing demographics and gait biomechanics, (b) quantify and visualize disease severity and individual biomechanical variability among asymptomatic and TKA candidates, (c) abstract decision rules to classify patients among the relevant patient severity phenotypes prior to arthroplasty, and (d) assess how patient phenotype prior to surgery relates to outcomes after arthroplasty. This work is being conducted in collaboration with the School of Biomedical Engineering at Dalhousie.
Acromegaly Facial Features – Novel Strategies Comparing Patients, Specialists And Computerized Facial Recognition Software
Collaborators: Syed Ali Imran (Dept. of Endocrinology, NSHA)
Synopsis: Acromegaly is a slowly developing disease due to high growth hormone levels released from a benign pituitary tumor that causes many complications such as severe facial disfigurement, enlargement of hands and feet, heart disease, cancer, sleep apnea, severe psychological problems and early death. In this project we aim to investigate a novel acromegaly screening strategy that asks non-specialists (particularly by patients themselves) to detect the presence of acromegaly using an acromegaly screening tool. We are developing a patient-centered home-based acromegaly screening mobile app that will enable patients to self-grade their photograph using the screening tool to detect the presence of acromegaly. We are investigating the use of facial recognition technology to grade the patient’s photograph to achieve automated acromegaly screening.
PLUS: Pathology Laboratory Utilization Scorecards
The ‘Choosing Wisely Canada’ movement is promoting sustainable healthcare by optimizing the utilization of healthcare services. Pathology laboratories provide service to help with disease diagnosis and therapeutic choices. A meta-analysis of 108 studies involving 1.6 million results from 46 of the 50 most commonly ordered lab tests, found that on average 30 % of all tests are likely to be unnecessary. The question pursued in this project is whether pathology test ordered in Nova Scotia are relevant and useful for patient management?
This project is in collaboration with Department of Pathology, and its objective is to optimize laboratory utilization from a primary care perspective by: (a) informing primary care physicians about their test ordering profile to help them reduce laboratory overutilization; (b) providing laboratory managers with intelligence about laboratory utilization patterns, to help them minimize waste.
In this health data analytics project, we are investigating and developing a digital health based Pathology Laboratory Utilization Scorecard (PLUS) system that leverages advanced data analytics and data visualization techniques to analyze ‘big’ laboratory data to generate objective, descriptive and predictive laboratory utilization insights to optimize laboratory utilization. We are applying advance data analytics methods to ‘learn’ physician-specific laboratory utilization models derived from their laboratory utilization data. To display laboratory utilization, we use visual analytics techniques to display an interactive and dynamic (i) physician scorecard for physicians to self-audit their test ordering patterns; and (ii) laboratory manager dashboard to illustrate laboratory utilization for resource planning. PLUS is a web-based system that after evaluation will be used to optimize the pathology laboratory in the central zone (Halifax) that annually performs on average 15 million laboratory tests for 200,000 patients.
Research Areas (Computer Science): Big data analytics, Machine learning, Data visualization, Dashboards, Data fusion
Research Areas (Health): Pathology, Choosing wisely, Health system use optimization
Project Duration: 2017 – 2019
SeDAn: SEmantics-based Data ANalytics Framework
This research is investigating plausible reasoning for semantic analytics over knowledge graphs, especially to analyze ‘big’ health datasets for question answering and hypothesis-testing.
Plausible Reasoning (PR) represents the plasticity element of human reasoning, which copes rather when confronted with incomplete data. In contrast to deductive reasoning, which reasons over a complete set of statements to infer a true statement, plausible reasoning can work with incomplete data as it derives plausible solutions by inferring the semantic relationships inherent in the data, thereby overcoming data completeness and somewhat correctness issues. Plausible reasoning is particularly akin to the clinical decision-making process of physicians, who consider the available patient information and use their tacit knowledge to infer a conclusion by discovering correlations to infer any missing information. Plausible reasoning mimics the physicians’ thinking process by inferring plausible solutions based on semantic relations inherent within the data.
The project has implemented a plausible reasoning framework—called SeDAn)—that leverages Semantic Web technologies for knowledge representation and reasoning, including the
Resource Description Framework (RDF), RDF Schema (RDFS), Web Ontology Language (OWL) and SPARQL to retrieve and manipulate the stored RDF data.
Plausible reasoning patterns developed in the project are used to answer medical questions retrieved from BioASQ challenges using standard clinical ontologies, DrugBank, Disease Ontology, and the semantic MEDLINE database as knowledge sources.
Research Areas (Computer Science): Semantic web, Knowledge graphs, Plausible reasoning, Query answering
Research Areas (Health): Health data analytics
Project Duration: 2015 – 2018
JADE: Juvenile idiopathic Arthritis Dialogue-based patient Education System
Juvenile Idiopathic Arthritis (JIA) is a life-long disease with outcomes that can include pain, prolonged use of medications, and disability. To manage their child’s condition, families often seek information to better understand the condition, therapy options, risk, and recovery trajectories. However, it is noted that families often feel overwhelmed by the volume, authenticity and format of the available patient information.
The JADE project is investigating and developing a novel, dialogue-based patient education approach that offers an interactive dialogue to discuss the issue with the user and in turn provide relevant information. In an interactive manner (human-like conversation), JADE allows the user to ask questions, seek clarifications about responses/findings, learn about the evidence supporting the response and seek alternative findings.
JADE uses an artificial intelligence based argument model to establish an interactive dialogue which uses a large volume of evidence-based digitized patient education material. JADE has implemented a comprehensive argument ontology, reasoning over the ontology allows the selection of the relevant educational material as per the discourse of the dialogue with the user who interact with JADE through a synchronous text-based dialogue interface. The interactive dialogue allows the user to explore a topic with respect to their own interests and apprehensions as opposed to being provided with a static, generic document. JADE is being evaluated at the Rheumatology Clinic at the IWK Health Centre (IWK), Halifax.
An Agile Semantic Web Platform for Knowledge-Centric Decision Support
Synopsis: This research program aims to advance semantic web methods to achieve agile knowledge management in terms of the delivering up-to-date, integrated and consistent knowledge to support knowledge-centric decision-making. The research program is investigating semantic web technologies to develop an agile knowledge management platform that offers: (a) Knowledge versioning mechanisms to incorporate new/updated knowledge to existing knowledge models (i.e. ontologies) to ensure that the modeled knowledge is current and consistent; and (b) Knowledge integration methods to integrate multiple knowledge models (i.e. ontologies) to yield a ‘complete’, contextualized and consistent knowledge model that can be reasoned over to provide decision support. The research methods are directed to the healthcare domain to: (i) dynamically adapt existing clinical guideline knowledge models to incorporate new or updated evidence/procedures as they become available; and (ii) integrate multiple CPG and clinical pathways to handle co-morbid diseases.
Duration: May 2013 – 2018
ADVICE (Agile Decisional Validation and Individualized Care Environment)
Synopsis: This multi-faceted research project is aiming to develop an innovative healthcare knowledge management infrastructure to support knowledge- and data-centric decision support services. ADVICE features four themes: (a) Transformation and translation of clinical guidelines to evidence-informed decision-support for care providers; (b) Engagement and empowerment of patients in their care process; (c) Health data analytics to derive health situational awareness; and (d) health knowledge formalization and mobilization. Semantic web based knowledge modeling and execution is central to the research program where we have developed a range of specialized and executable ontologies to model clinical guidelines, behavior models and clinical workflows that are incorporated within dedicated decision support systems. The project objectives are: (a) To provide effective healthcare knowledge to health professionals where and when they need it to help them make evidence-informed, safe and cost-effective care decisions, and (b) To assist patients through a continuum of personalized, proactive and persistent home-based care services throughout their care journey.
Duration: January 2012 – December 2017
IMPACT-AF (Integrated Management Program Advancing Community Treatment of Atrial Fibrillation)
Synopsis: This research program offers an innovative clinical decision support environment to support both healthcare providers and patients in the management of Atrial Fibrillation (AF) at the primary care level. IMPACT-AF is a knowledge management infra-structure to: (a) computerize the Canadian clinical guidelines on the diagnosis and management of AF; (b) offer decision support services to recommend patient-specific, guideline-driven diagnostic and therapeutic interventions; (c) deliver proactive alerts and reminders to flag potential adverse trends in the patient’s evolving condition; and (iii) assist patients to self-manage AF in a home-based setting. The IMPACT-AF will be accessible through mobile devices and web-based applications. IMPACT-AF will be deployed across 200 primary care clinics in Nova Scotia and will be used to study over 4000 patients.
Funding Agency: Bayer Inc.
Duration: January 2013 – December 2017
H-DRIVE (Healthcare Data Retrieval, Inferencing and Visualization Environment)
Synopsis: This project aims to derive actionable intelligence and situational awareness from health data to help improve healthcare services through predictive planning, resource optimization, evidence-based decision-making and personalized medicine. H-DRIVE is developing a range of big data management and analysis services that will enable end-users to perform complex and contextualized healthcare analytics. The innovation of H-DRIVE is that it will place the power of ‘big’ data healthcare analytics directly in the hands of the end-users to: (i) self-investigate contextualized operational and research questions using their own datasets, (ii) self-generate customized, consumable and clear insights with respect to their current needs, and (iii) self-display and communicate those insights through interactive visualizations. The project is conducting research in the area of semantics and big data, especially investigating semantic data analytics.
Duration: January 2013 – December 2016
D-WISE: An E-health Solution Operationalizing Behaviour Change Models and Clinical Guidelines for Patient-centered Diabetes Self-management
Synopsis: This project is inspired by CIHR’s Strategy for Patient Oriented Research, where it aims to assist patients in modifying their behavior towards healthy lifestyles and adherence to therapeutic programs. This objective is achieved by: (a) assisting family physicians to administer behavior change strategies to their patients with chronic diseases, and (b) empowering patients to modify their behavior through personalized behavior modification programs. D-WISE (Diabetes Web-Centric Information and Support Environment) aims to influence positive behavior change in type-2 diabetic patients by (i) translation of clinical guidelines and behavior change models through web-based clinical decision support tools to assist family physicians and diabetes educators to offer behavior change strategies to diabetes patients; and (b) the engagement of patients to self-manage their diabetes aided by personalized behavior change and disease management strategies that are delivered and monitored via a mobile app.
Funding Agency: CIHR (Catalyst Grant)
Duration: March 2013 – February 2014
TIME (Tools, Information, Motivation, Environment) for health
Synopsis: In this project, we have proposed and implemented a two-level information personalization scheme that integrates both health and behavior determinants to personalize self-management programs for healthy eating. A semantic web based framework computerizes the constructs of social cognition theory to achieve behavior modification through personalized messages that are customized to the individual’s self-compliance challenges and healthy eating targets. A mobile TIME app has been developed that assists families to achieve healthy eating targets by enacting the personalization strategy that deliver timely alerts and reminders to individuals.
Funding Agency: Heart and Stroke Foundation of Canada & CIHR
Duration: January 2011 – January 2014
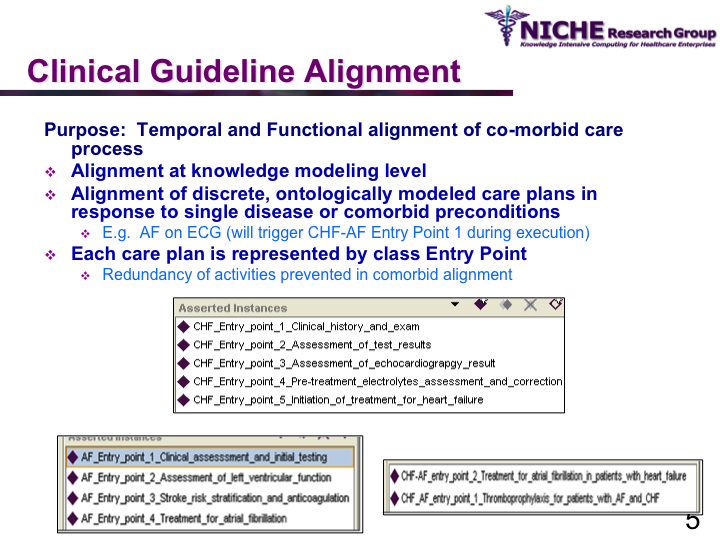
COMET (Co-morbidity Ontological Modeling & ExecuTion) Framework
Synopsis: This research program aims to deliver clinical guideline based recommendations for managing co-morbid conditions via clinical decision support systems. The research involves the systematic and safe synthesis of multiple disease-specific clinical guidelines to manage co-morbid situations. Taking a knowledge management methodology, COMET offers a semantic web based framework for the (a) computerization of clinical guidelines as an executable OWL ontology, (b) synthesis of ontologically-modeled clinical guidelines, using ontology alignment mechanisms, to respond to comorbid situations; and (c) execution of the aligned clinical guidelines using reasoning engines to deliver patient-specific recommendations to manage co-morbidities. COMET provides an interactive web-based clinical decision support system for family physicians to handle Chronic Heart Failure (CHF), Atrial Fibrillation (AF) and co-morbid CHF and AF. COMET has been evaluated through a pilot study involving family physicians for its usability and utility in handling complex chronic disease cases at the primary care level.
Funding Agency: Bayer Inc.
Duration: July 2008 – September 2013
YouCan (An Integrated Knowledge Management Framework for Supporting Young Cancer Survivors)
Synopsis: This research project aims to address the educational, communication and care management aspects of youth cancer care programs in order to provide quality, safe, responsive and standardized care to youth cancer survivors aged 12-29. Using semantic web based technologies, we have computerized shared-decision making constructs and the clinical guideline on follow-up of youth cancer survivors to develop a decision support system that assists healthcare providers to provide youth cancer survivors with a personalized self-management diary based on their specific disease condition and care needs, to help them safely manage their condition in a home-based setting. The YouCan Ontology represents over 600 decision rules relating to 110 drug side-effect interactions for 22 different therapeutic agents, which upon execution based on the patient’s profile recommend range of information and action items, packaged as a personalized YouCan Diary for youth cancer survivors.
Funding Agency: CIHR (Catalyst Grant)
Duration: May 2010 – March 2013
Semantic Web Framework for Developing Personalized Information and Adaptive Web Services
Synopsis: Knowledge Management
Funding Agency: NSERC Discovery Grant
Duration: April 2007 – March 2013
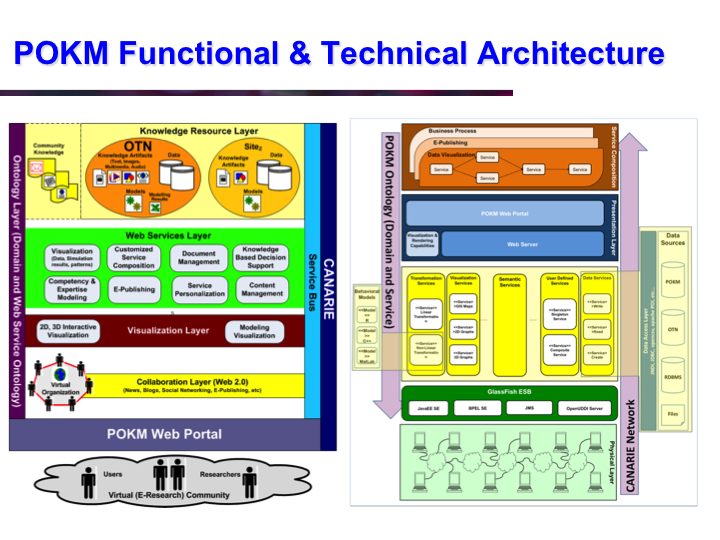
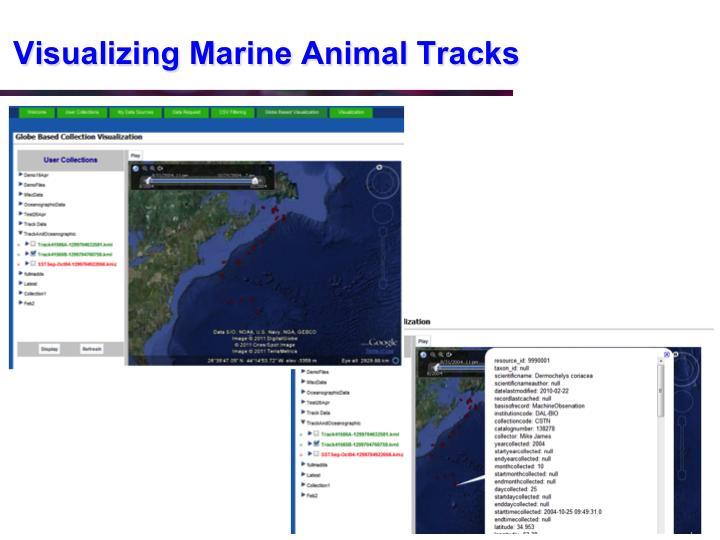
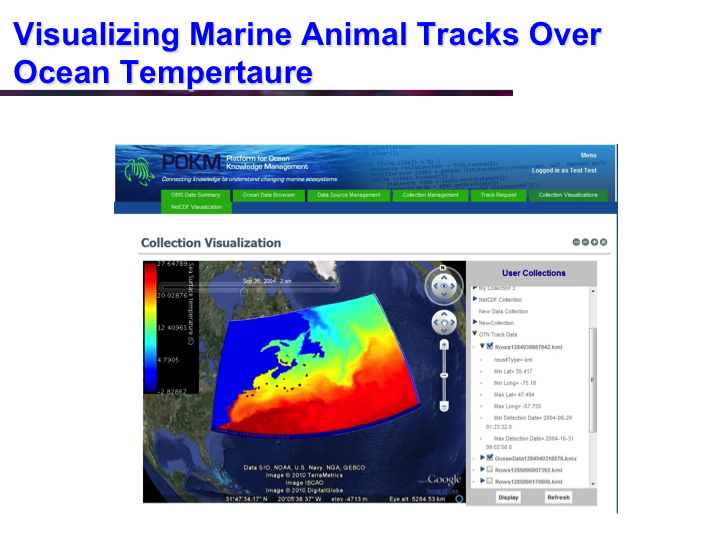
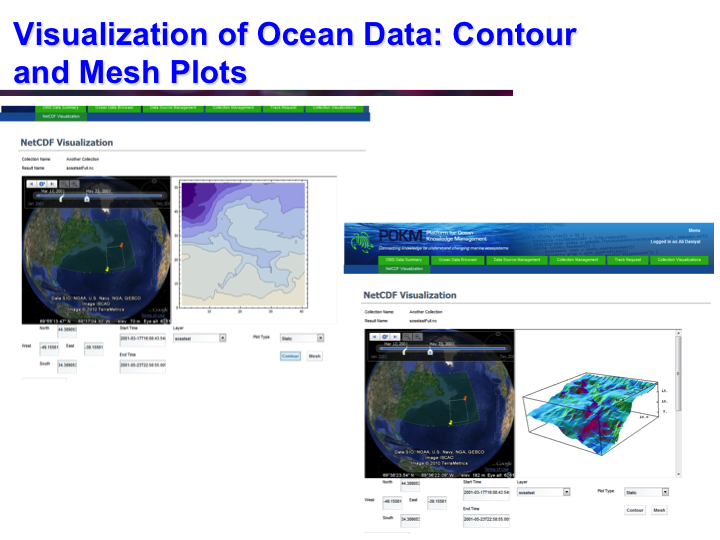
POKM: Platform For Ocean Knowledge Management
Synopsis: POKM offers an e-Science platform featuring specialized scientific data management services to assist scientists to: (a) collect and share multi-modal ocean data; (b) perform analytics and simulations; (c) visualize multiple data layers in different formats; (d) publish simulation models; and (e) design and execute complex experiments by composing specialized experimental workflows. The technical architecture of POKM showcases a unique synergy of semantic web, services oriented architectures and web services. At the knowledge level, POKM offers a high-level abstraction of the scientific domains through domain-specific ontologies to establish inter-discipline interoperability for data integration and sharing. POKM offers a range of e-Science services that are accessible through a web-based portal. POKM leverages the CANARIE network to transfer of high-volumes of ocean data. The project was conducted in collaboration with the Ocean Tracking Network (OTN) at Dalhousie University. View Images
Funding Agency: CANARIE
Duration: March 2009 – March 2012
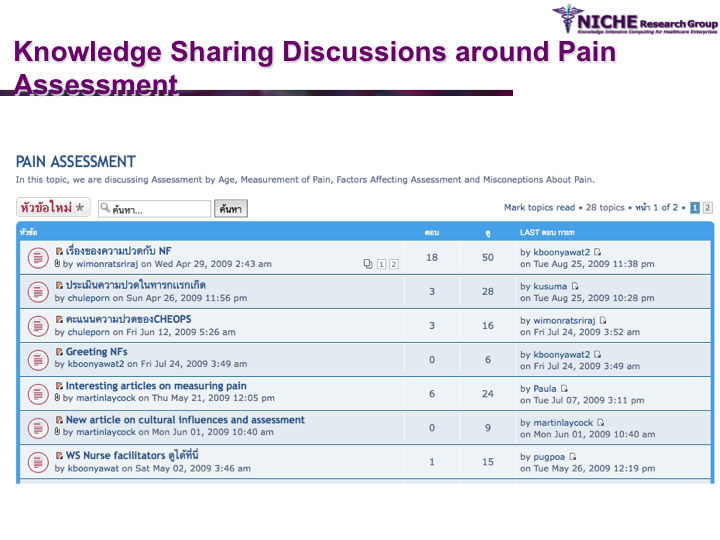
Paediatric Pain Management in Urban and Rural Thailand
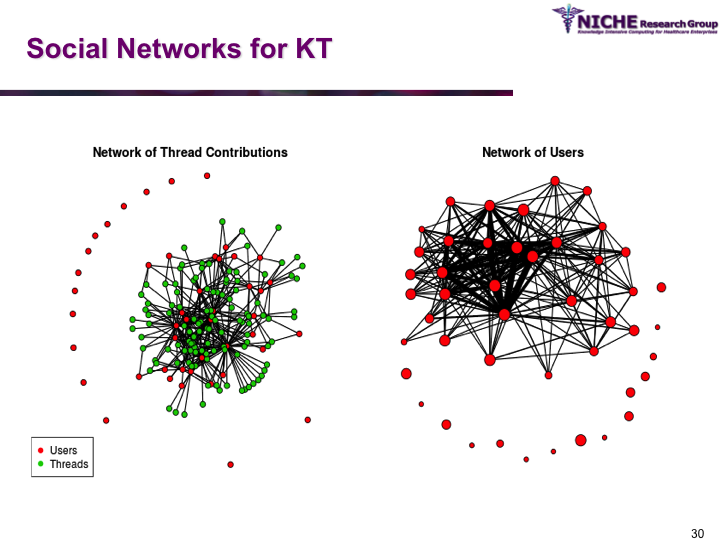
Synopsis: Pediatric Pain Knowledge Translation Framework: This knowledge translation project was conducted in Thailand. The project featured the design and implementation of an innovative technology-driven knowledge translation strategy targeting pediatric pain management in 8 hospitals in the Isan province (northeast Thailand). The knowledge translation tool was a specialized web 2.0 based discussion forum that allowed the sharing of both experiential and evidence-based knowledge amongst a community of around 60 pediatric pain practitioners. Social network analysis and discussion content analysis allowed better understanding of knowledge translation pattrens and practices in terms of the knowledge sharing behaviors, discussion foci and collaboration determinations of the local pediatric pain practice community. The knowledge translation strategy provided opportunities to formulate regional clinical guidelines on pediatric pain management.
Funding Agency: Teasdale-Corti Global Health Research Partner Program, International Development Research Centre (IDRC), Canada
Duration: June 2006 – May 2012
Cruise Control: First Gear—Development of the Community Research Alliance
Synopsis: Health Informatics
Funding Agency: NSHRF
Duration: May 2010 – April 2011
CarePlan: Personalized Lifelong Patient Care
Synopsis: Knowledge Management and Health Informatics
Funding Agency: Agfa Healthcare, Canada
Duration: February 2005 – January 2010
Knowledge Management Solutions for Practimax™ EHR System
Synopsis: Knowledge Management & Health Informatics
Funding Agency: Nova Scotia Govt.
Duration: February 2009 – May 2009
Surveillance Strategies for Identifying Adverse Drug Events: Lessons From Public Health
Synopsis: Knowledge Management
Funding Agency: Canadian Institute for Health Research (CIHR)
Duration: January 2008 – June 2009
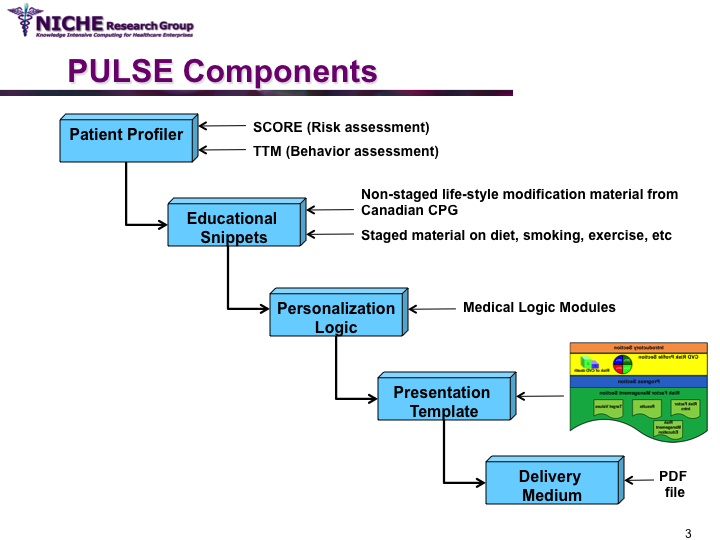
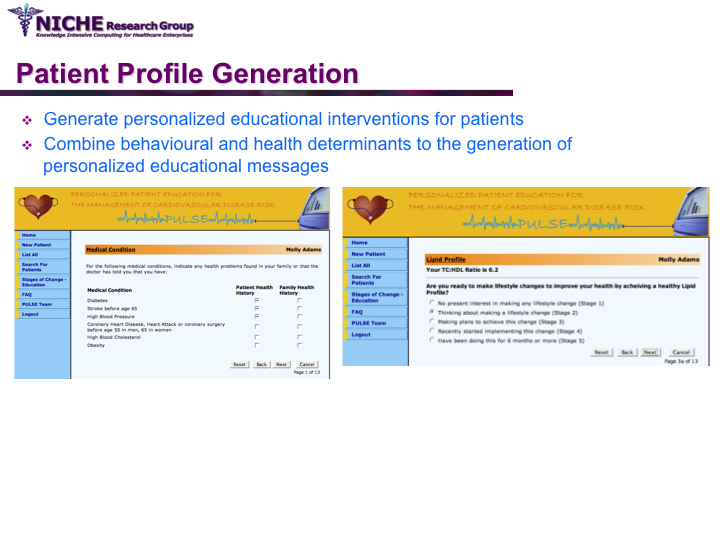
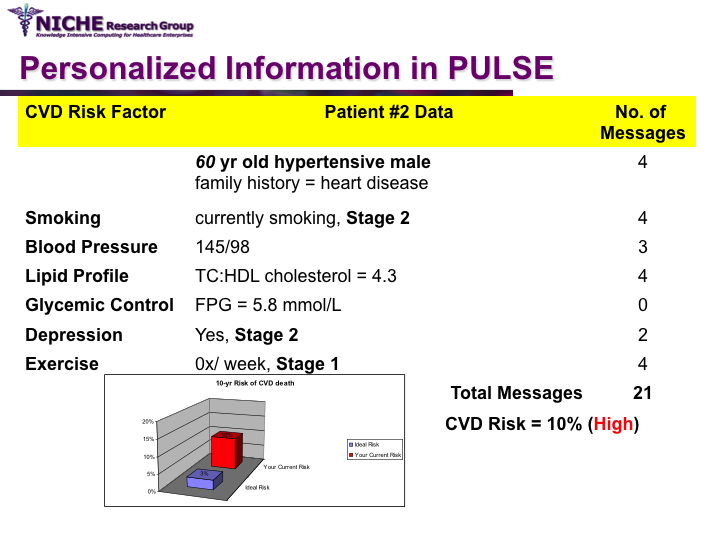
PULSE: Personalization Using Linkages of SCORE and behaviour change readiness to web-based Education
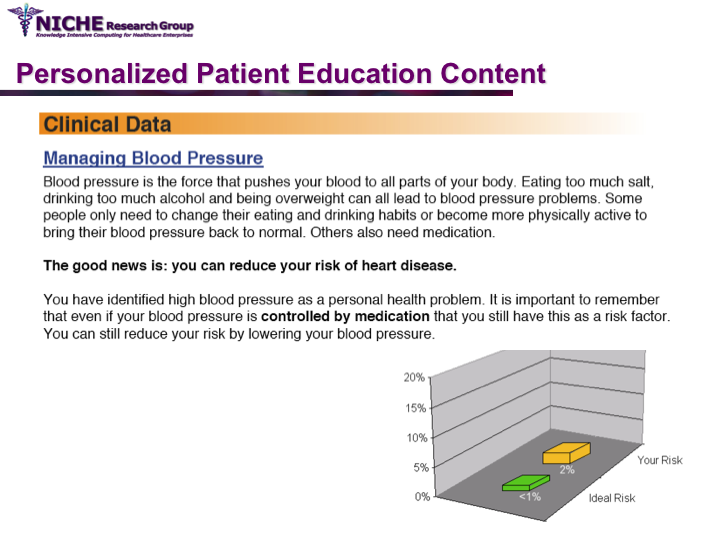
Synopsis: This project featured a compositional information personalization strategy that personalizes patient educational content by dynamically selecting and combining relevant ‘snippets’ of information items. The personalization approach was applied within the PULSE system to generate personalized cardiovascular disease risk management educational package. The key feature of PULSE is the determination of the patient profile that integrates the patient’s (a) behaviorial readiness to positive lifestyle assessed using the using the Transtheoretical Model, and (b) current health determinants and care needs. PULSE is accessible to users through a web application and it has been successfully evaluated through a pilot user-study involving CVD patients.
Duration: January 2005 – December 2008
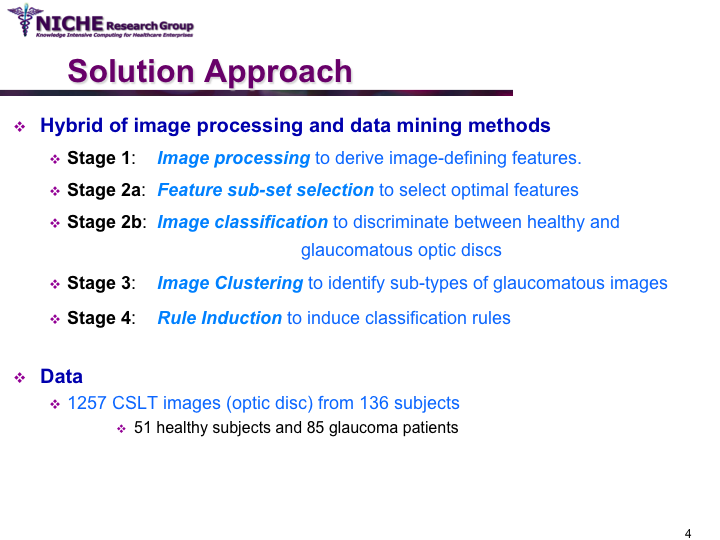
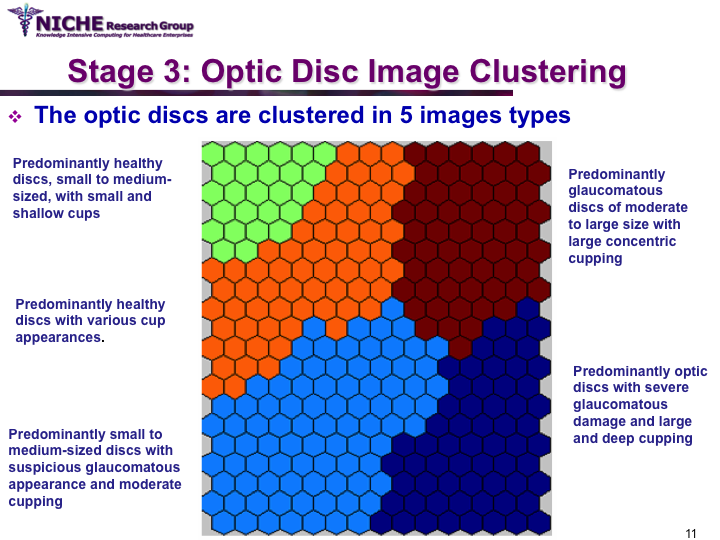
Data Mining Approach For Optic Nerve Analysis to Detect Glaucoma
Synopsis: This project developed an automated approach to analyze Confocal Scanning Laser Topography (CSLT) images of the optic nerve to support the diagnosis of glaucoma. Taking a data mining approach, we developed a glaucoma decision support system that offers: (a) the classification of the optic disc images to distinguish between healthy and diseased optic discs; and (b) the identification of the sub-types of glaucomatous optic disc damage. The decision support system offered the functionality to (i) diagnose the presence of glaucoma and the type of glaucoma; (ii) visualize the temporal progression of glaucoma within a patient; and (iii) identify noisy images that can then be discarded from diagnostic tasks. View Images
Funding Agency: Nova Scotia Health Research Foundation (NSHRF)
Duration: October 2003 – September 2008
Standardization of Prostate Cancer Management
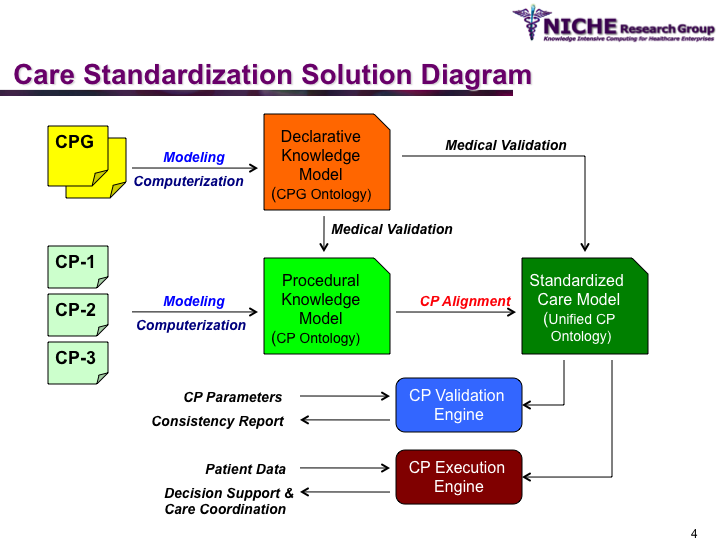
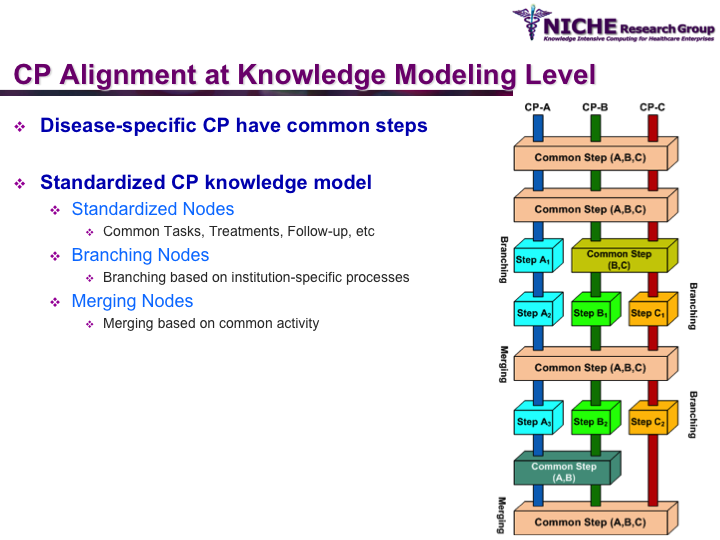
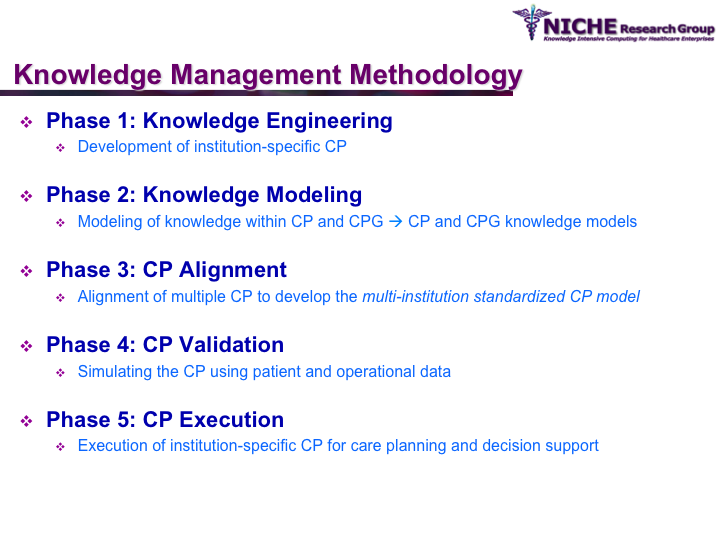
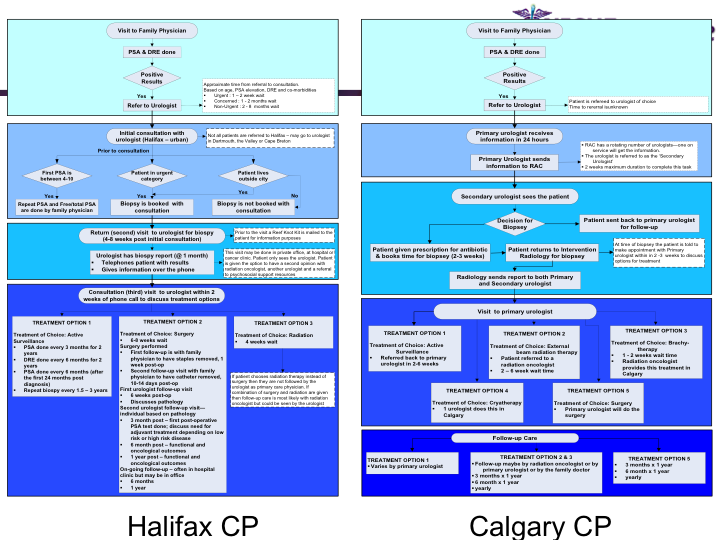
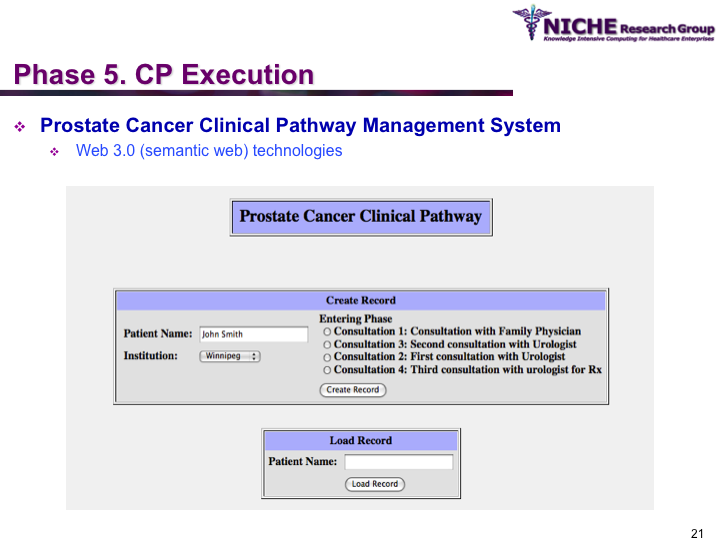
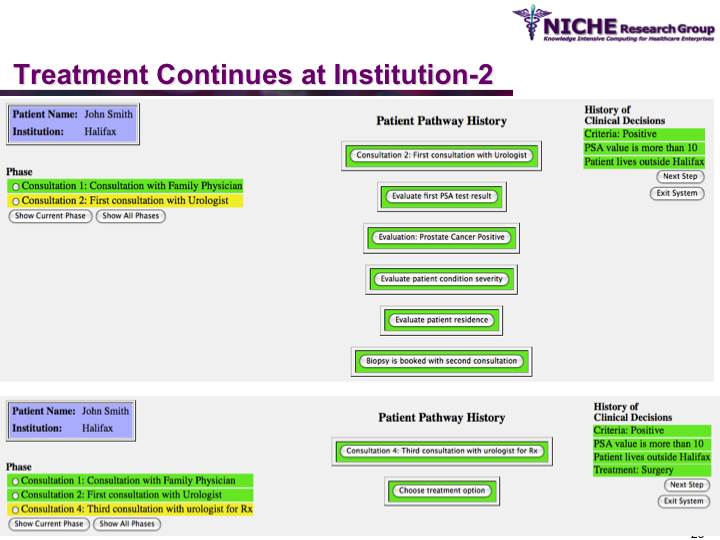
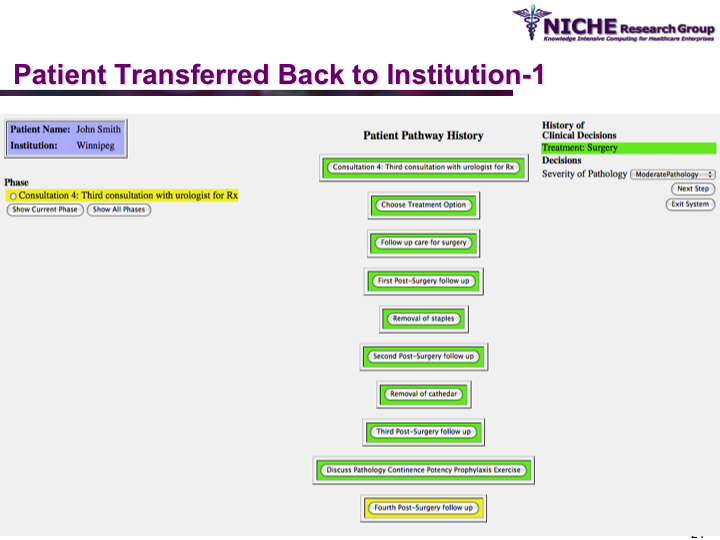
Synopsis: This research program pursues the standardization of specialized care for a specific condition—in this case prostate cancer—across multiple institutions. Given that each institution has a unique Clinical Pathway (CP) for managing a condition, this project aims to generate a standardized condition-specific CP by aligning the independent CPs along common care activities/processes/policies/outcomes. Taking a semantic web based knowledge modeling approach, care standardization is achieved by: (a) computerizing institution-specific CPs using a standard CP ontological model; (b) aligning the different ontologically-modeled CP to generate a standardized CP; and (c) executing the standardized CP, using patient-data and provider input, through a web-based decision support system. As a pilot study, we developed a prostate cancer management system that models and aligns the CP of three Canadian hospitals, thus streamlining the transfer of prostate cancer patients across different institutions based on a standardized CP.
Funding Agency: National Cancer Institute, Canada
Duration: March 2007 – December 2007
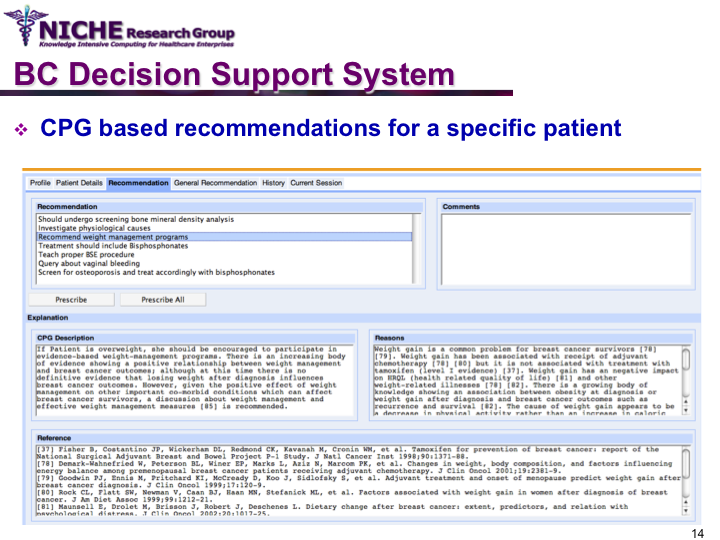
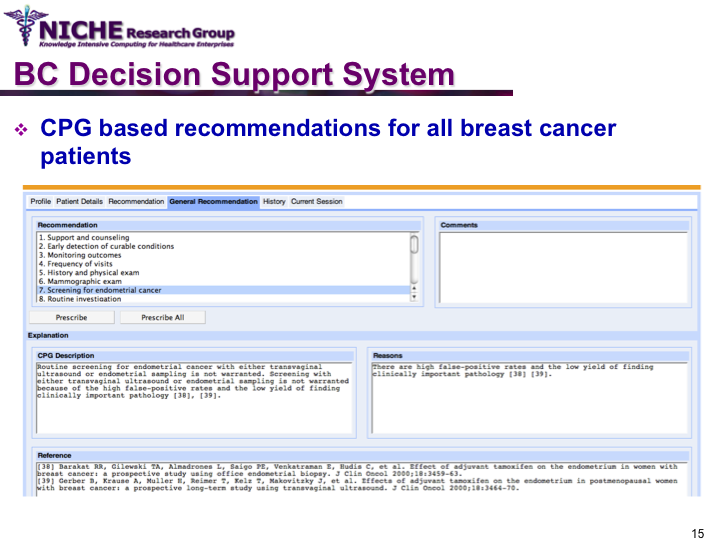
Clinical Guideline Based Breast Cancer Follow-up Decision Support System
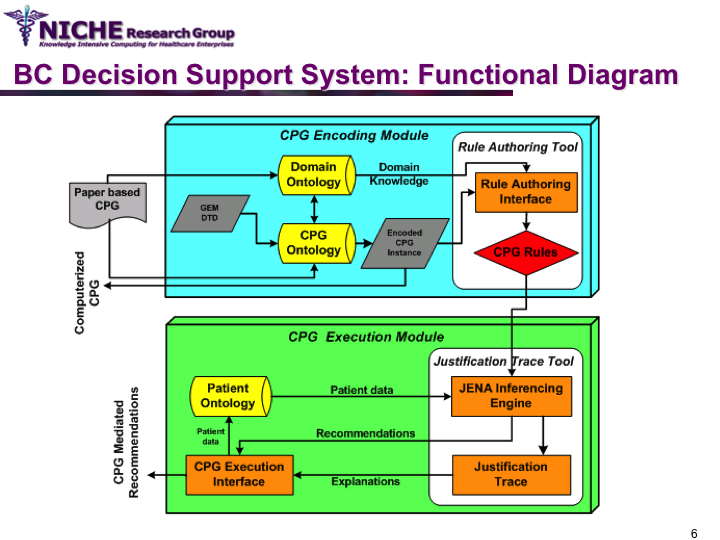
Synopsis: This knowledge translation project enabled the implementation of the Canadian clinical guidelines for the follow-up care of Breast Cancer (BC) to support family physicians to deliver breast cancer follow-up care and patient education. Taking a semantic web approach, the BC guideline was modeled as an ontology which was executed using a logic-based execution engine to provide to ‘trusted’ guideline-driven recommendations. The project yielded a web-based decision support system that allowed family physicians to (a) access and utilizes the BC clinical guideline at the point of care to provide standardized follow-up care; and (b) offer customized patient educational information targeting disease management, lifestyle behaviors and psychosocial support.
Funding Agency: Nova Scotia Health Research Foundation (NSHRF)
Duration: September 2005 – September 2007
Concept Mapping and Semantic Modelling on Peer-to-Peer Networks
Synopsis: Knowledge Management
Funding Agency: NSERC (Collaborative Research and Development Grants)
Duration: September 2004 – September 2007
A Knowledge Management Framework for Tacit Knowledge Acquisition, Sharing and Operationalization
Synopsis: Knowledge Management
Funding Agency: NSERC (National Science & Engineering Research Council) Discovery Grant
Duration: July 2003 – June 2007
Scientific Visualization through Data Mining of Electronic Commerce & Health Informatics Large Scale Data Sets
Synopsis: Health Informatics & Data Mining
Funding Agency: Canadian Foundation for Innovation (CFI)